Introduction
The purpose of this post is to evaluate the structure of the training data used in running the S2AND algorithm; understanding the file formats will help us insert our own, unique data for author disambiguation
Folder Structure
Here is what the full data directory looks like:
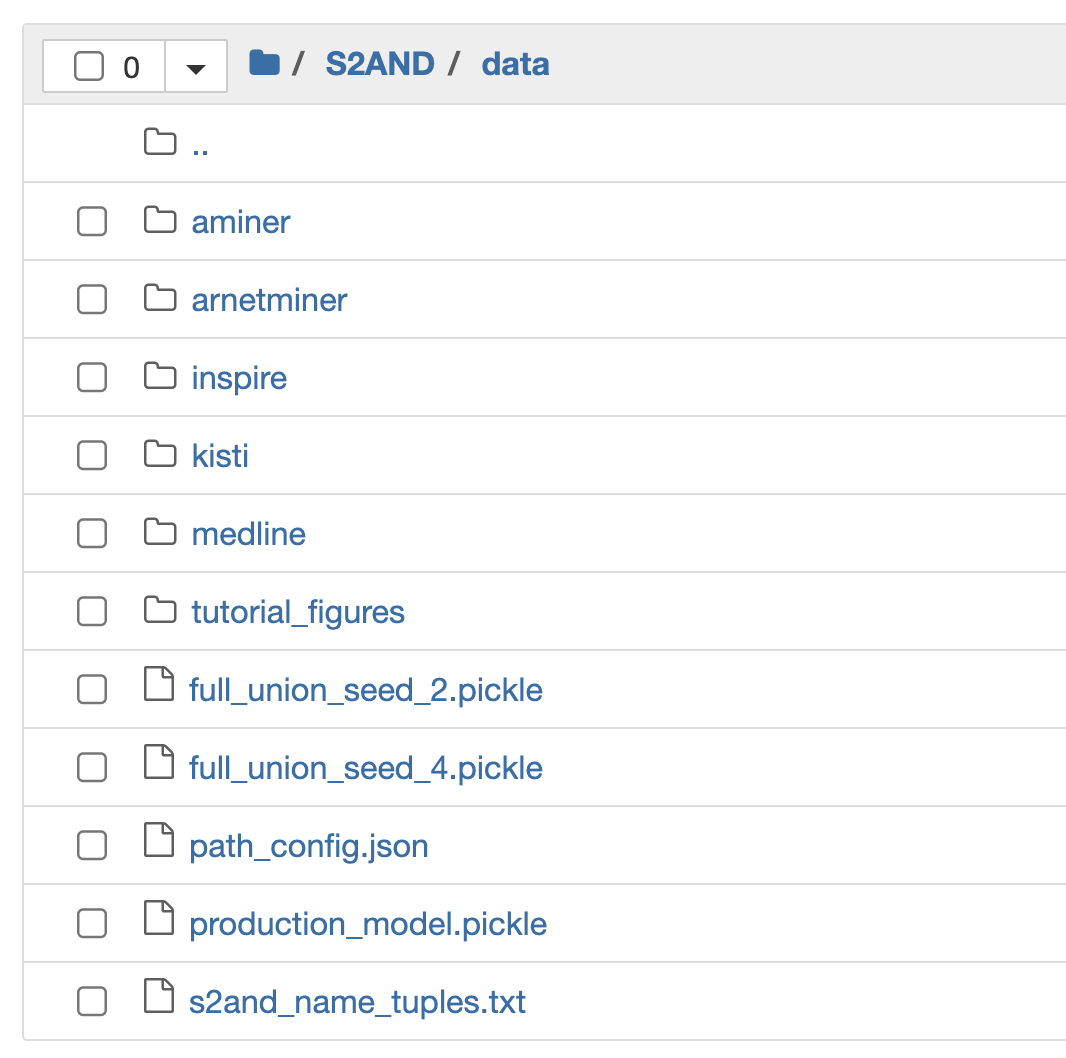
As you can see, the S2AND repo provides five folders for five different test runs of the algorithm, namely:
- Aminer
- ArnetMiner
- Inspire
- Kisti
- Medline
We also have access to the production-level, pretrained S2AND model that’s ready for us to plug in and use:
- production_model.pickle
The path_config.json is simply a config file to tell S2AND the absolute path of our data folders (make sure to update this!)
Frankly speaking, I’m not too sure what the following files are for, but that’s okay for now, because they are not needed for this post!
- full union seed _2.pickle
- full union _seed_4.pickle
- s2and_name_tuples.txt
Input Data
Here is what the file structure looks like for one of the datasets (ArnetMiner). Note that this structure is similar across all the remaining folders.
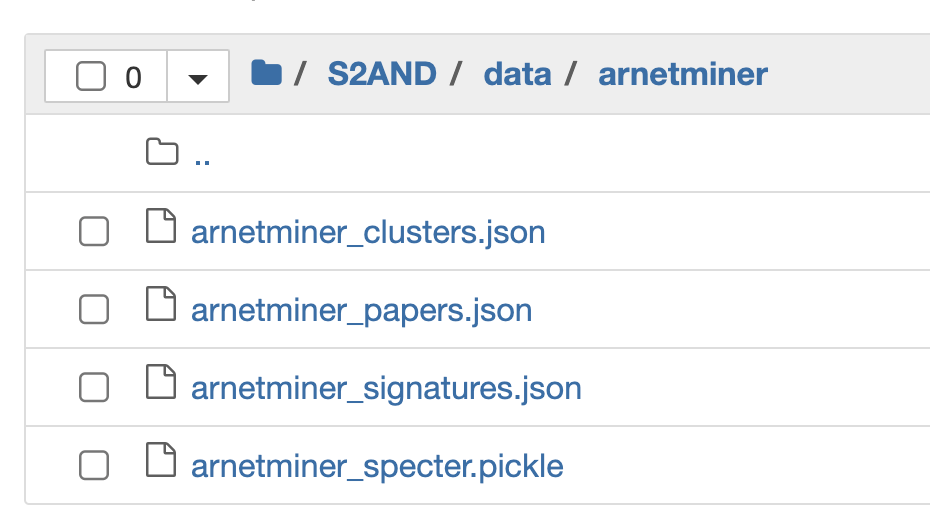
The 3 main input files that we will feed into our model are:
- arnetminer_papers.json
- arnetminer_signatures.json
- arnetminer_specter.pickle
Note: arnetminer_clusters.json is the end result file that’s generated after running the S2AND algorithm (we will revisit this shortly).
Papers.json
Here is what one record looks like from the papers.json file. For this snippet, keep track of the paper_id field.
"15718873": {
"paper_id": 15718873,
"title": "A predictive empirical model for pricing and resource allocation decisions",
"abstract": "We present a semi-parametric model that describes pricing behaviors in a market environment, and we show how that model can be used to guide resource allocation and pricing decisions in an autonomous trading agent. We validate our model by presenting experimental results obtained in the Trading Agent Competition for Supply Chain Management.",
"journal_name": null,
"venue": "ICEC",
"year": 2007,
"references": [
40571,
556512,
670266,
1058249,
1937296,
40571,
556512,
151245949,
167908624,
208846619
],
"authors": [
{
"position": 3,
"author_name": "Paul R. Schrater"
},
{
"position": 2,
"author_name": "Maria L. Gini"
},
{
"position": 1,
"author_name": "John Collins"
},
{
"position": 4,
"author_name": "Alok Gupta"
},
{
"position": 0,
"author_name": "Wolfgang Ketter"
}
]
}
Signatures.json
Here are what two records looks like from the signatures.json file. Notice how these two in particular have the same paper_id; these are the separate instances for each author from the previous papers record with the same paper_id.
Despite sharing the same paper_id, these two records differ in a few other attributes. Specifically, both records have different signature_id values; this is the unique author-paper combination. Moreover, we find that the position value within the author_info field reflects the same position value from the papers.json file.
"34": {
"author_id": 6879414,
"paper_id": 15718873,
"signature_id": "34",
"author_info": {
"given_block": "a gupta",
"block": "a gupta",
"position": 4,
"first": "Alok",
"middle": null,
"last": "Gupta",
"suffix": null,
"affiliations": [
"University of Minnesota"
],
"email": null
}
},
"3059": {
"author_id": 1225211537,
"paper_id": 15718873,
"signature_id": "3059",
"author_info": {
"given_block": "j collins",
"block": "j collins",
"position": 1,
"first": "John",
"middle": null,
"last": "Collins",
"suffix": null,
"affiliations": [
"University of Minnesota"
],
"email": null
}
}
One thing to note: author_id can be duplicated for each record. This is because author_id is based on the author’s name (first, middle, and last)–any author with the same name has the same author_id but different signature_id.
This is worth clarifying once more:
- signature_id: Unique author name + paper combination
- author_id: Unique author name combination
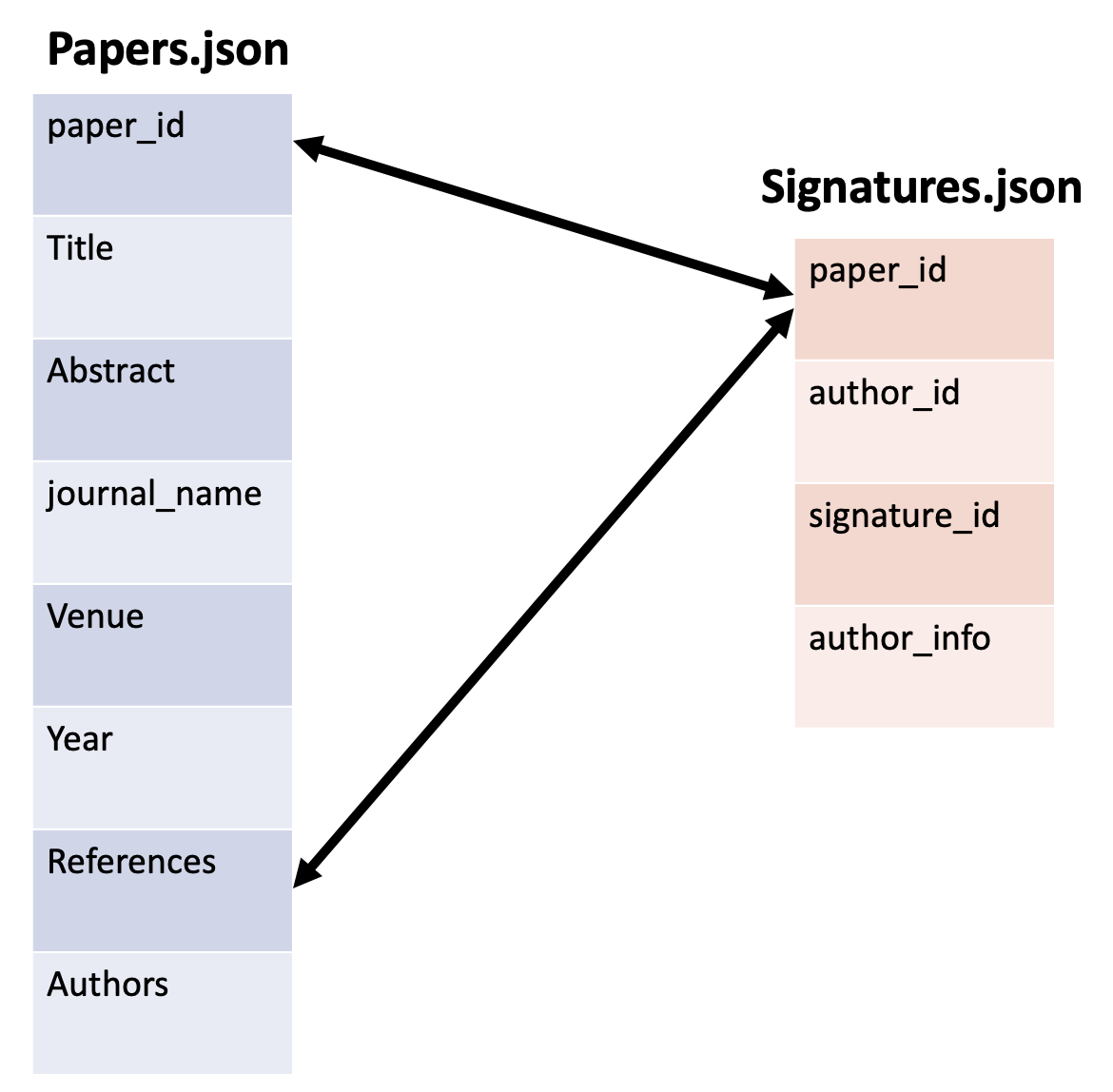
Relationship between Papers.json and Signatures.json
The common variable between both of these datasets is quite simple: paper_id serves as a common key.
Specter.pickle
The last input file is the specter embeddings.
The specter.pickle is a pickle file that, when loaded into the python environment, is a tuple of two elements.
We may load the file by using the following snippet:
import pickle
# Open the pickle file for reading
with open('data/arnetminer/arnetminer_specter.pickle', 'rb') as f:
# Load the pickled data into a variable
data = pickle.load(f)
The structure of this tuple is as follows:
- data[0][n] reveals the (768,) dimensional numpy.ndarray for the n-th record
- data[1][n] reveals the paper id for the n-th record
For this particular dataset, the embeddings are based on the concatenation of the title and abstract; when we insert our own data, we may choose to replicate this process, or only embedd one over the other.
Here is what the spectral embeddings pickle file (specter.pickle) looks like for one paper:
array([-3.55426264e+00, -3.36183834e+00, -9.27550435e-01, 1.05726647e+00,
-4.33013964e+00, -2.57947731e+00, 9.08111572e-01, -2.62764573e-01,
2.07375789e+00, -2.22142792e+00, 5.78262031e-01, 2.92265147e-01,
...
6.44651127e+00, -4.98596787e-01, -5.25885010e+00, 4.34468699e+00,
2.59104633e+00, 2.19220567e+00, -3.41159296e+00, 4.48349762e+00,
2.82836962e+00, 2.65981579e+00, 2.97524214e+00, -3.73605371e+00,
-1.00283372e+00, 1.84344888e+00, -4.66459274e+00, 1.65630072e-01,
8.57315636e+00, 2.66784430e-03, -1.72782099e+00, -3.47309875e+00])
Pretty cool, huh?
Ground Truth Data
clusters.json
The goal of S2AND is to determine which set of signature_idss represent the same author.
Some of the folders contain the ground truth for comparison; here is what the final clusters.json looks like:
"AM_ajay gupta_1": {
"cluster_id": "AM_ajay gupta_1",
"signature_ids": [
"0",
"1",
"4",
"13",
"16",
"18",
"23",
"26"
],
"model_version": -1
},
"AM_ajay gupta_8": {
"cluster_id": "AM_ajay gupta_8",
"signature_ids": [
"2",
"3",
"5",
"6",
"10",
"11",
"12",
"15",
"17",
"19",
"25",
"28"
],
"model_version": -1
},
"AM_ajay gupta_7": {
"cluster_id": "AM_ajay gupta_7",
"signature_ids": [
"7",
"8",
"9",
"21",
"22",
"24",
"30"
],
"model_version": -1
},
"AM_s. huang_5": {
"cluster_id": "AM_s. huang_5",
"signature_ids": [
"5215"
],
"model_version": -1
},
As we can see, the specific blocking method will vary, but the end result is the same: unique groupings for each set of signature_ids
Generating our own data
For this step, we will need to generate two files to get started: (1) papers.json (2) signatures.json.
While we may scrape the web for author information, for the purposes of this demo, we may instead choose to create synthetic data that illustrates the same point.
Using ChatGPT
Using ChatGPT, I was able to generate the two files.
I first started with papers.json, since signatures.json would be based off of the authors presented in the papers.
I prompted my query to ChatGPT by provided the details of what the fields in the papers.json would appear, and also providing one example record from the S2AND datasets.
You can view the full ChatGPT chat log here as a text file.
Once I had a set of 6 records for the papers.json, it was now time to create the signatures.json file.
Since signatures.json file is simply a unique author-paper combo, which is based off of papers.json, we can easily create a function that parses through the author names, collects the meta data (names, position, paper_id), and automatically create out blocking schema.
Here’s what the function looks like:
import random
def generate_signatures():
with open('papers.json') as f:
papers = json.load(f)
signatures = {}
author_id_set = set()
for paper_id, paper in papers.items():
for author in paper['authors']:
author_id = random.randint(1000000, 9999999)
while author_id in author_id_set:
author_id = random.randint(1000000, 9999999)
author_id_set.add(author_id)
signature_id = len(signatures)
given_block = author['author_name'].split()[0][0].lower()
last_name = author['author_name'].split()[-1].lower()
block = given_block + ' ' + last_name
author_info = {
'given_block': given_block + ' ' + last_name,
'block': block,
'position': author['position'],
'first': author['author_name'].split()[0],
'middle': ' '.join(author['author_name'].split()[1:-1]) or None,
'last': last_name,
'suffix': None,
'affiliations': [],
'email': None
}
signatures[str(signature_id)] = {
'author_id': author_id,
'paper_id': int(paper_id),
'signature_id': str(signature_id),
'author_info': author_info
}
with open('signatures.json', 'w') as f:
json.dump(signatures, f, indent=4)
return 2
As you can see, this function creates the signatures.json for us, and since this is a synthetic dataset for demonstration purposes, we can fill in the additional fields (email and affiliation) as we please.