Introduction
The purpose of this post is to provide readers a quick recap on the S2AND paper by Allen AI. Readers are encouraged to review the original paper for technical details they wish to further investigate. Additionally, Daniel King and Sergey Feldman have done a great job documenting S2AND development in a separate Medium post by Sergey Feldman.
For a post demonstrating the implementation of S2AND, feel free to read the second part of this post.
Introduction
What problem does this algorithm solve?
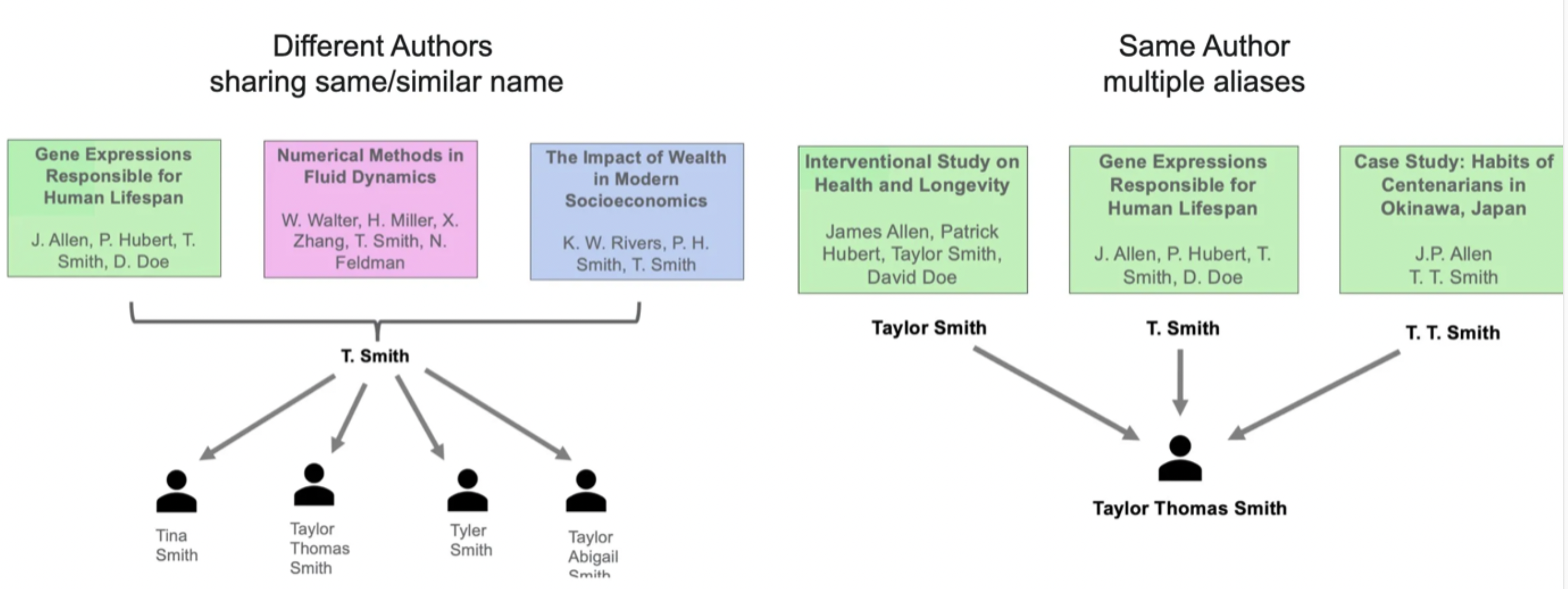
S2AND is an algorithm to resolve author disambiguation in publication papers when an ORCID (or some unique identifier) is not present. These scenarios include:
- Authors with similar names
- Authors with multiple aliases
- Changes in author properties (e.g. institution, name, etc.)
What is the output from the S2AND algorithm?
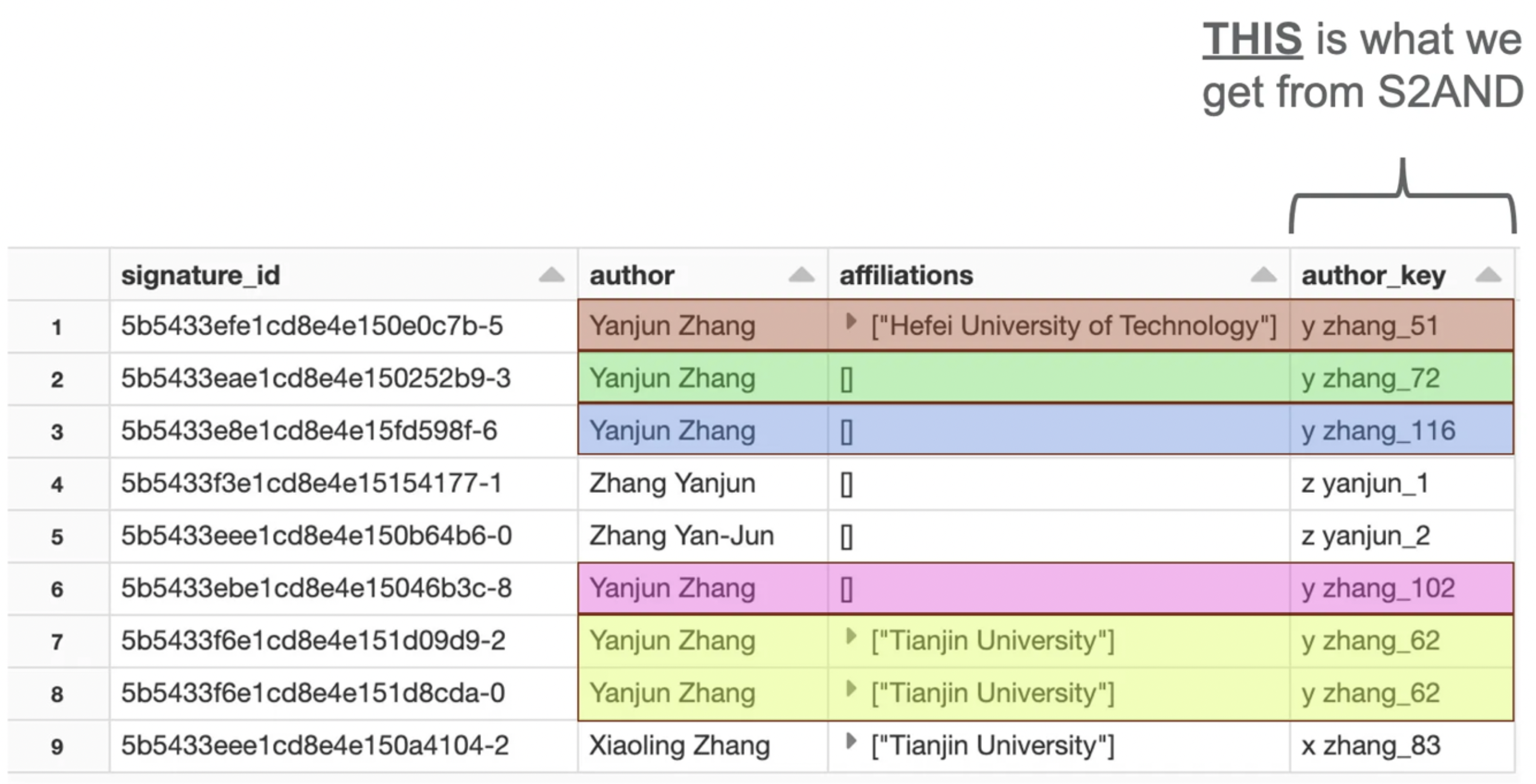
The final output from S2AND is a unique author key for each record (author-paper combination). This is essentially adding a column to the records that serves as an ORCID.
What does this paper offer?
For the community of ML practitioners, there are two main contributions from this paper:
- S2AND, a new, unified training dataset and evaluation benchmark for author name disambiguation (AND)
- A new, state-of-the-art open-sourced AND algorithm
Additionally, the paper itself provides additional insight, such as
- Experiments showing that training on S2AND improves generalization
- A comparison against the existing Semantic Scholar production system
About the Data
Why the need for a new AND dataset?
A common issue with many pre-existing AND datasets is that they tend to cover idiosyncratic and biased slices of the literature. For instance, the AMiner dataset consists of only Chinese names, while SCAD-zbMATH contains only mathematics papers.
Matters are further complicated when each dataset contains unique features not present in other datasets.
Such niche datasets may impair generalization performance; algorithms trained to perform well on one on dataset may generalize poorly to others.
What makes the S2AND dataset different?
S2AND attempts to ameliorate the previously mentioned issues by harmonizing eight disparate AND datasets into a uniform format with a consistent, rich feature set drawn from the Semantic Scholar (S2) database.
What datasets does S2AND consist of?
- Aminer
- ArnetMiner
- INSPIRE
- KISTI
- Medline
- PubMed
- QIAN
- SCAD-zbMATH
About the Model
How does the S2AND algorithm work?
Broadly speaking, there are three steps in the algorithm:
- Group candidate records into disjoint, potentially-coreferent blocks
- Score the similarity of each pair of records within a block based on available features
- Cluster the records based on the pairwise scores
How are records grouped into blocks?
This is often performed heuristically, based on author names; the goal is to group together records such that each pair of records within a block potentially refers to the same author (potentially-coreferent).
A record (AKA, a signature) is simply any author-paper combination.


The blocks are disjoint, meaning that no two blocks can contain the same record (author-paper combination).

Although different datasets use different blocking functions, a typical choice is to put all records with the same last name and first initial into the same block.
How are records scored for similarity?
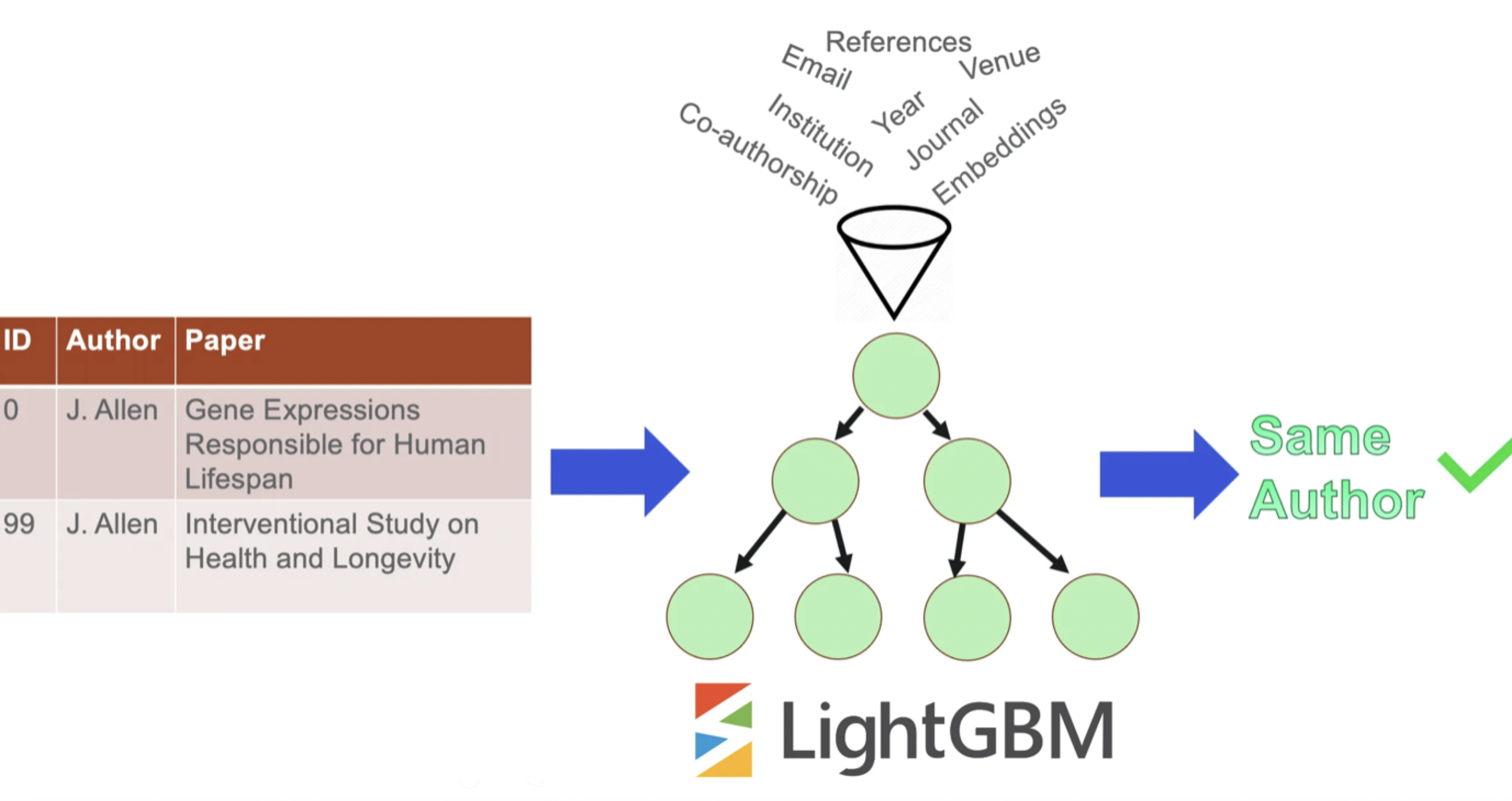
Within each block, similarity of each record pair is estimated using Gradient Boosted Trees (LightGBM); if the score is high enough, we assume that the two records were written by the same author.
It is important to note that S2AND uses an ensemble of two classifiers:
- A classifier trained on the full feature set
- An identical classifier trained on “nameless” feature set
This “nameless” feature set is identical in every respect to the full feature set, but does not contain any features related to the author names (co-author names are still included).
What clustering method does S2AND use?
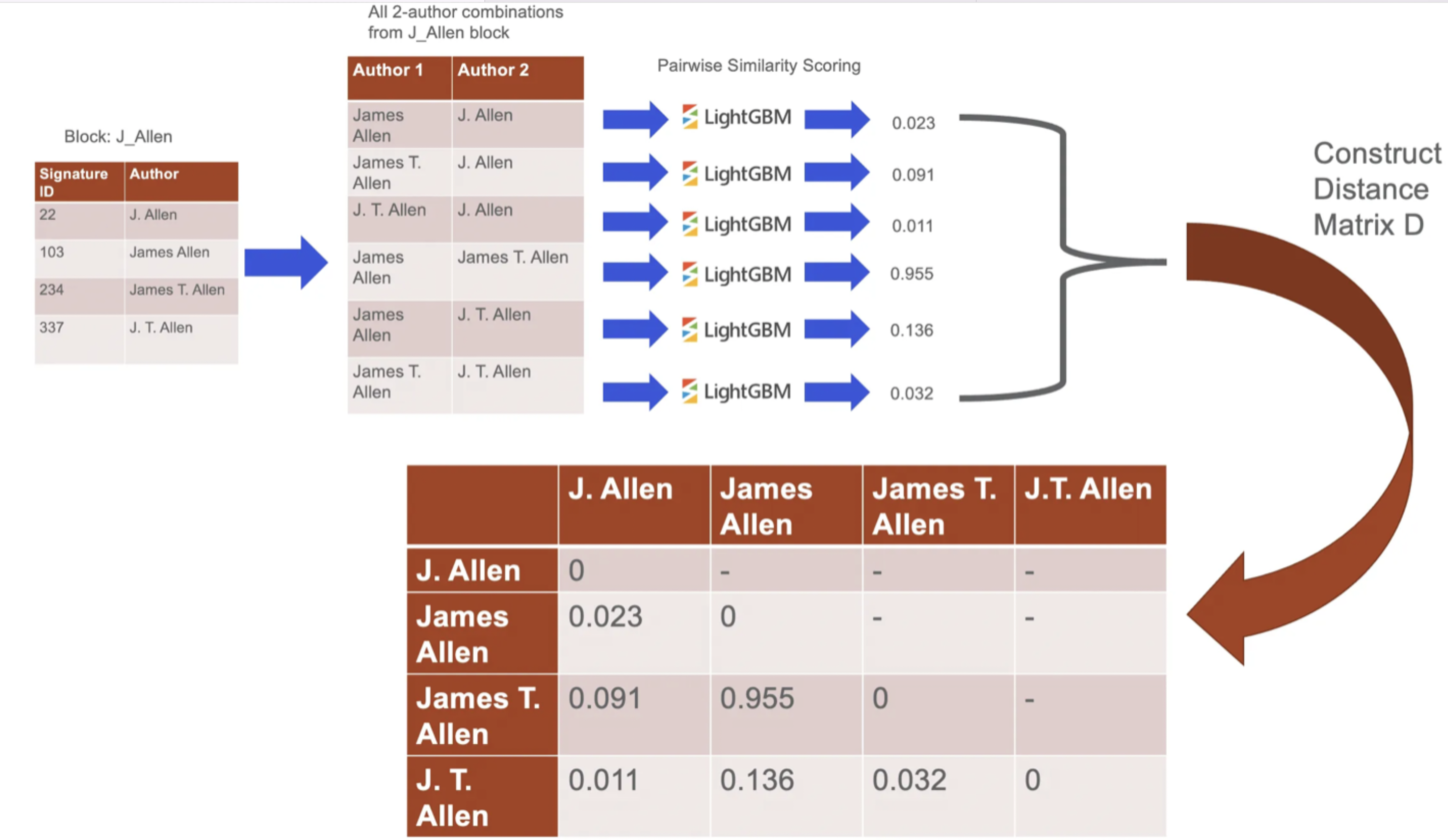
After having scored each pair within a block, S2AND implements Hierarchical Agglomerative Clustering (HAC).
Specifically, the classifier from the previous step is used to construct a distance matrix D where Dᵢⱼ is the probability that two records i and j are not by the same author. Each block is then partitioned into clusters with HAC over the matrix D.
The paper mentions how the clustering will depend on a linkage function that estimates the dissimilarity between two clusters (in terms of the pairwise distances between the individual elements of each cluster); experiments suggest that a straightforward average of all the pairwise distances performs best.
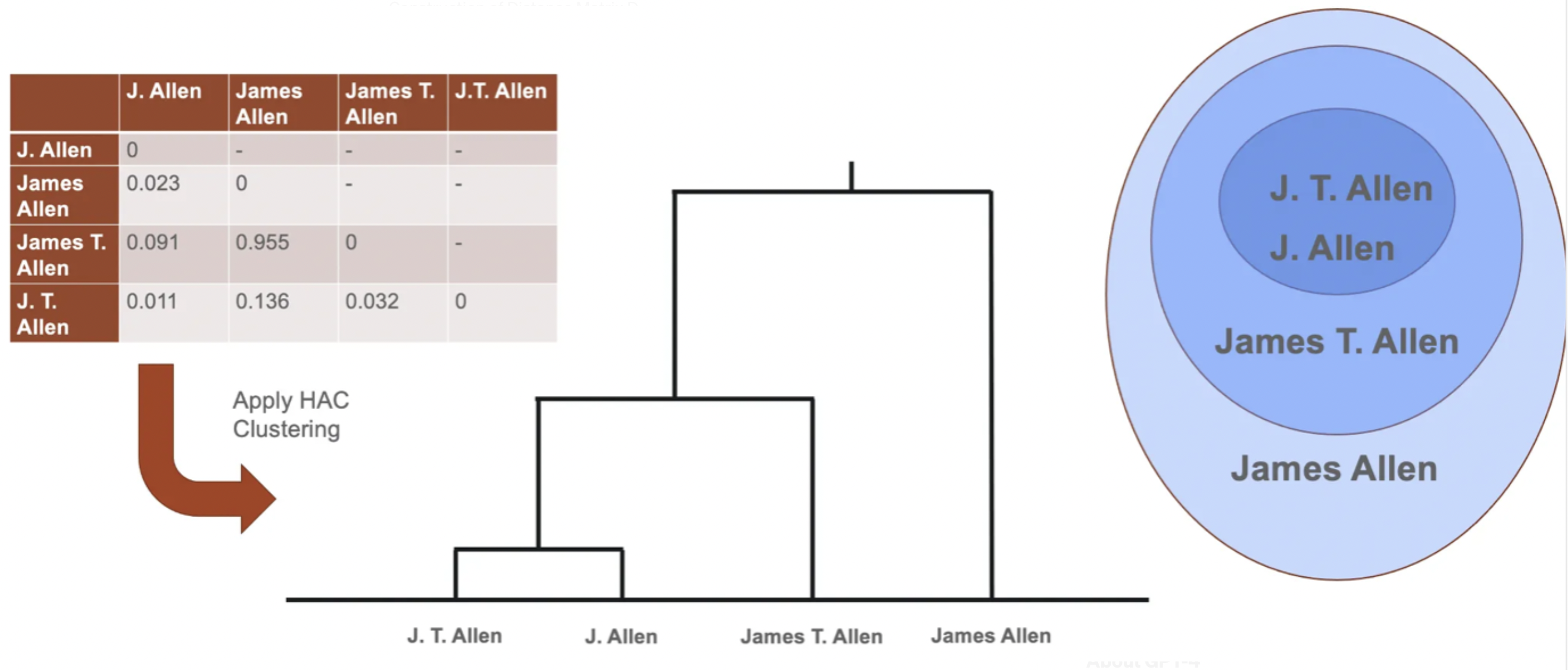
Note: The use of HAC was considered after comparing only two methods: DBSCAN and HAC. The paper highlights the use of other clustering methods for future studies.
About the Features
What features are the models trained on?
The paper uses a total of 15 features to estimate similarity between documents. A list of these features with a brief description is shown below.

While many of these features are self-explanatory, a few warrant further explanation.
“Name counts” reflects name popularity, which is estimated as number of distinct referents of author names across S2.
“SPECTER embeddings” are the vectorized representations of the paper title and abstract (concatenated as one text). SPECTER refers to a specific document-embedding algorithm trained on the citation graph to produce paper embeddings applicable across multiple tasks.
What are the most important features?
Feature importance is estimated using SHapley Additive exPlanations (SHAP).
The results indicate that three features appear to consistently rank as most important across all datasets (no order):
- SPECTER embedding
- Affiliations
- Name counts (last names)
Although these features appear vital in AND, this does not imply these three features alone would led to adequate model training. This is because the authors also compared feature variations (i.e. excluding certain features to evaluate performance) and found that most design alternatives hurt performance.
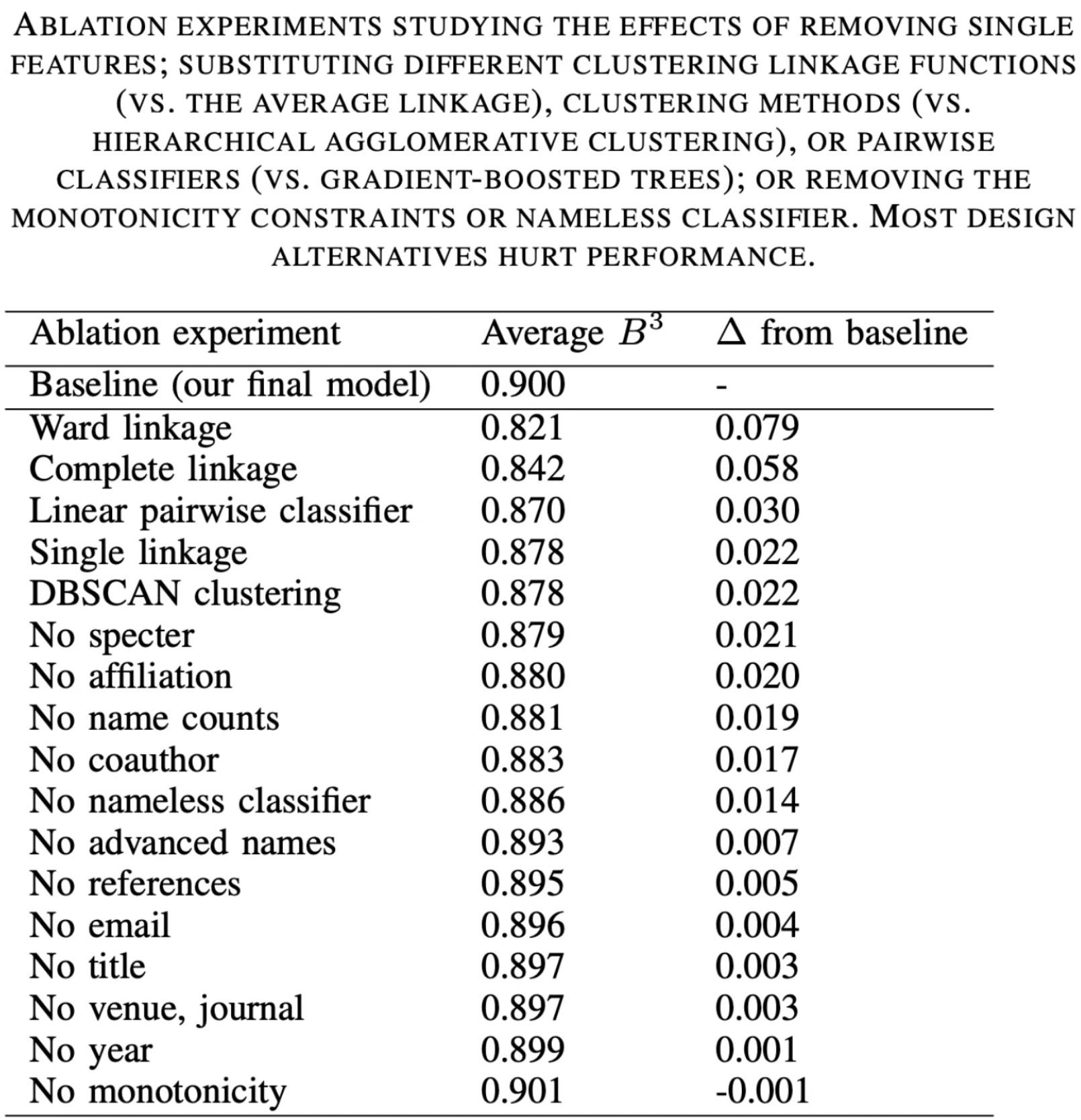
In other words, it is best practice to train with the full set of features.
What additional features are helpful?
In addition to the original feature list, the authors found that imposing a feature-wise monotonicity constraints led to substantial qualitative improvements in B³.
The monotonicity constraints influence the features in order to:
- Regularize the model
- Constrain the model to behave sensibly when faced with data from outside the training distribution
This is often best explained through example:
Holding all other features constant, the model decreases the output probability if the year difference between two records increases.
Note: the output probability is the probability that any two records from the same block are by the same author.
About Using S2AND
What is the main takeaway when training with S2AND?
Training on the union of all datasets, rather than any single data source, is an effective way to transfer to out-of-domain data and produces models that are more robust across all the existing datasets.
What are the limitations of S2AND?
- S2AND features, particularly SPECTER embeddings, are only intended for English-language records
- If certain metadata is missing, its absence could inadvertently encourage the model to learn spurious interactions and make counterintuitive predictions
- Although the model is resistant to obvious errors (e.g. blocking “John” and “James” together because they share the same initial), this is not to suggest that all errors are accounted for, as authors can change their names, and names can be mis-extracted from papers
- At the time of this post, there is no successful solution for recovering from name blocking errors
- Lossy transliteration of names may occur (e.g. Chinese names to English)
- The HAC pipeline does not allow for the similarity of one pair of records to influence the similarity of another pair of records
