Introduction
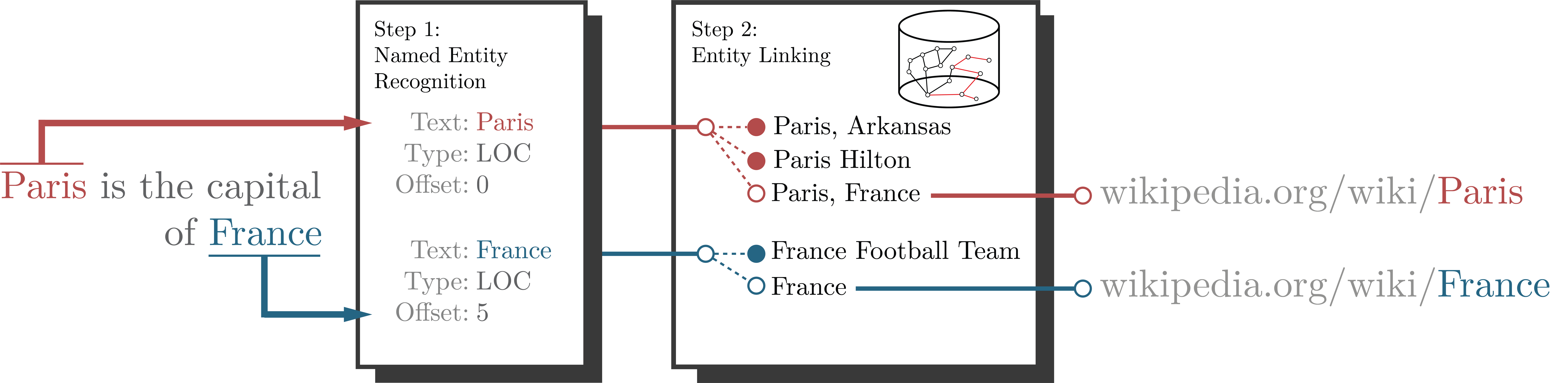
Entity linking is a critical task in natural language processing that involves linking entities mentioned in text to their corresponding entries in a knowledge base.
In this project, we implement a creative approach to entity linking that leverages the complementary capabilities of Semantic Role Labeling (SRL), Named Entity Recognition (NER), and BERT-based FAISS indexing.
We use SRL and NER to extract relevant queries from the input text, which are then used to perform a search in our FAISS index.
The index is built using a BERT-based synonym model that generates embedding vectors for each entity in the knowledge base. The candidate entities are then ranked using nearest neighbors of embedding space.
Our proposed approach to entity linking has significant implications in the medical space for patient care and biomedical use cases by improving the accuracy and efficiency of clinical decision support systems, as well as facilitating the extraction of valuable insights from biomedical literature.
The code, data, and instructions for this algorithm can be found in this repo
Algorithm Overview
Here’s how this entity-linking algorithm works:
- Generate pre-embeddings of database terms
- Parse sentence for query candidates
- Search for nearest neighbors of query, and obtain top K best matches
Details
Generating Pre-Embeddings
This step involves using FAISS indexing for similarity search, as well as a transformer model for computing the embeddings.
Hugging Face provides excellent support and documentation on how to use FAISS sematic search within the transformers library.
The tutorial that Hugging Face provides is practical because we will also need to use a transformer model to generate the embeddings for each of the terms in our database.
Having the right embedding matters plays VITAL role in search results–so much so, that choosing the correct model is the make-or-break point for the entire algorithm.
Fortunately, I did not have to try out too many models before arriving at tekraj/avodamed-synonym-generator1.
Interestingly enough, because this was a biomedical use case for entity-linking, I figured a model like michiyasunaga/BioLinkBERT-large would be fantastic–but performance was far worse!
That’s when my mentor Sutanay Choudhury famously told me the relation between vector similarity and synonym similarity:
“closeness in vector space does not imply synonym similarity”
Parsing sentences for query candidates
In this step, we parse each sentence of a given paragraph/context to extract query candidates; we call them queries because they are the search inputs that we eventually feed into the FAISS similarity search.
The goal of this parsing is to extract medical terms, signs, and symptoms related to what the patient is experiencing.
To achieve this goal, we use two parsing methods:
- Named Entity Recognition (NER)
- Semantic Role Labeling (SRL)
We use these two methods together because NER often works well to extract commonly recognized words, while SRL works to provided an extra boost in extracting words/phrases outside of what NER can detect.
For example, given the sentence,
The patient is suffering from a raging headache. He mentioned this past week that he has fever, nausea, and vomiting. He has trouble concentrating and often feels anxious.
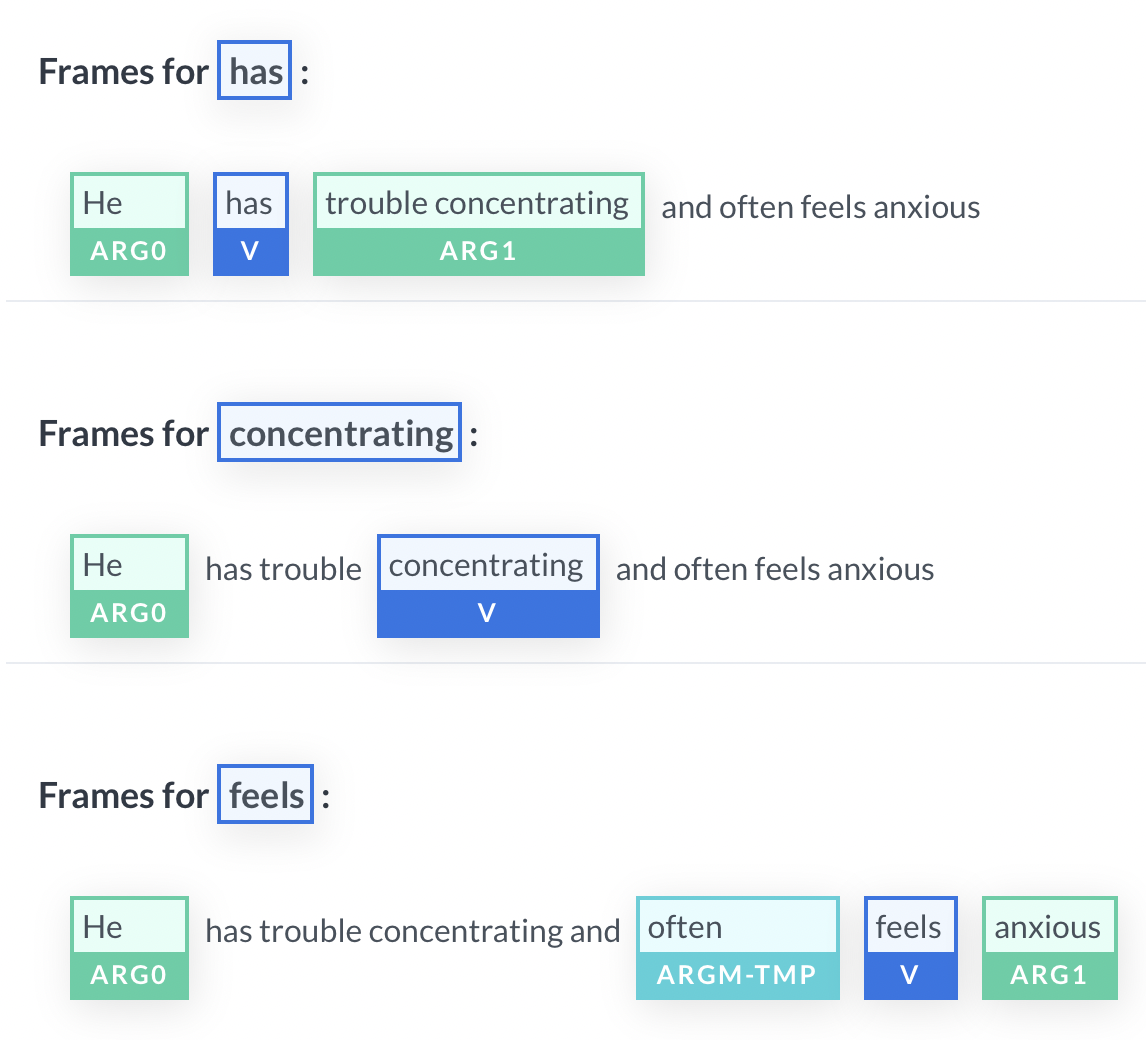
A biochemically pretrained NER model would work great at picking up words like “fever”, “nausea”, “headache”, and “vomiting”, but would have a harder time picking up “trouble concentrating” or “anxious”–that’s exactly where SRL helps to scoop up the extract helpful and descriptive terms.
The specific models I’ve found to work well for this project are scispaCy’s en_ner_bc5cdr_md model for NER, and Allen AI’s SRL BERT model (2020.12.15) for SRL (direct model download link here)
NOTE: Allen AI also has a web demo for the SRL model that anyone can try here.
Similarity Search via nearest neighbors
Using the saved pre-embedding file that we computed earlier, we now begin to embedd each query using our same huggingface model, and compute the embedding distance between that query and all of the embedded terms from the saved file.
Conclusion
Overall, the results are pretty good from a search standpoint; download the repo and give it a try!